

11/12/2025
ABSTRACT
This study aimed to isolate, qualitatively screen, and identify azo dye–degrading microorganisms from the activated sludge of a textile wastewater treatment system. Congo Red (100 mg/L) was used as the sole selective dye in Minimal Salt Medium to enrich potential degraders. A total of ten bacterial isolates (M1–M10) were successfully obtained, all exhibiting decolorization halos ranging from 22 to 46 mm on MSM agar. Among them, isolates M1 and M8 demonstrated the strongest activity, forming clear zones of 46 mm and 44 mm after 48 hours, respectively. Molecular identification through 16S rRNA gene sequencing revealed that M1 shared 99.87% similarity with Bacillus subtilis, while M8 shared 99.86% similarity with Bacillus licheniformis. Both species are known for their production of extracellular oxidative enzymes, such as laccases and peroxidases, which play key roles in azo bond cleavage and aromatic amine detoxification. These findings highlight the presence of native, dye-adapted Bacillus strains with strong decolorization capacity in textile wastewater sludge and provide foundational data for developing bioaugmentation agents applicable in sustainable textile effluent treatment systems.
Keywords: Activated sludge; Azo dye decolorization; Bacillus licheniformis; Bacillus subtilis; Textile wastewater.
1. INTRODUCTION
The textile dyeing industry generates substantial volumes of wastewater characterized by pronounced fluctuations in pH, temperature, and organic load, along with the significant presence of azo dyes, which account for 60–70% of the dyes used worldwide [1]. These compounds contain the highly stable azo bond –N=N–, making them resistant to conventional biodegradation and contributing to reduced photosynthetic activity in aquatic ecosystems as well as visual pollution [2]. More critically, the incomplete degradation of azo dyes can release aromatic amines with high toxicity, many of which are known to be mutagenic and carcinogenic, as reported for the degradation products of Congo Red [3]. This poses an urgent demand for treatment technologies that are both effective and environmentally sustainable.
Although physical and chemical treatment processes can rapidly remove color, their high operating costs, substantial production of secondary sludge, and limited mineralization efficiency hinder their practical applicability [4]. In contrast, biological treatment has emerged as a promising approach due to its low cost, operational simplicity, and environmental compatibility. Numerous studies have demonstrated the azo dye–degrading potential of bacteria and fungi isolated from contaminated environments. Lysinibacillus sphaericus and Aeromonas hydrophila have been reported to remove 88–91% of Joyfix Red within 72 hours [5], while Absidia spinosa M15 achieved 88% removal of Cresol Red through a combined mechanism of biosorption and biodegradation [6]. Several Aspergillus species have also shown high removal efficiency for Direct Violet, reaching up to 93% through extracellular enzymatic activity [7]. The genus Bacillus continues to receive considerable attention due to its dual ability to decolorize dyes and reduce COD, as exemplified by Bacillus aryabhattai [8].
However, most existing studies rely on exogenous or pure laboratory strains, whereas indigenous microorganisms residing in activated sludge from textile wastewater treatment plants are naturally adapted to high dye concentrations. This represents a critical research gap, as native strains often exhibit superior degradation performance and greater applicability for developing practical bioremediation agents. Rapid screening of potential strains using halo zone formation on agar plates provides an effective preliminary assessment of azo dye degradation before species identification via 16S rRNA gene sequencing, the standard method for determining taxonomic classification and predicting functional potential [9].
Based on these scientific foundations, the present study aims to isolate and qualitatively screen indigenous microorganisms from activated sludge in a textile dyeing wastewater treatment system to identify strains with the highest Congo Red degradation capability. Selected strains are then subjected to 16S rRNA gene sequencing for accurate taxonomic identification, providing essential groundwork for subsequent studies on optimizing azo dye degradation conditions
2. MATERIALS AND METHODS
2.1. Sample source and culture media
Microbial samples were collected from the activated sludge of the textile wastewater treatment system at Phong Phu Corporation. Samples were maintained under sterile conditions at 4 °C and processed within 24 hours after collection.
The chemicals used in this study included the azo dye Congo Red (Sigma) and the components of the Minimal Salt Medium (MSM), namely Na₂HPO₄, KH₂PO₄, NH₄Cl, NaCl, MgSO₄, CaCl₂, FeSO₄·7H₂O. Nutrient Agar (NA) was used for culture maintenance.
2.2. Isolation and qualitative screening of dye-degrading microorganisms
2.2.1 Isolation procedure
Isolation was performed through an enrichment culture technique. Ten grams of activated sludge were suspended in 100 mL of MSM supplemented with Congo Red at a concentration of 100 mg/L. The mixture was incubated in a shaker incubator at 35 °C and 150 rpm for 24 hours.
After incubation, 10 mL of the enriched suspension was transferred to 90 mL of fresh MSM containing Congo Red (100 mg/L), and this subculturing step was repeated three times to selectively enrich microorganisms capable of decolorizing the dye.
From the final enrichment culture, serial dilutions were prepared and spread-plated onto MSM agar containing Congo Red. Colonies exhibiting clear decolorization halos were selected and repeatedly streaked on NA plates to obtain pure isolates.
2.2.2 Qualitative assessment of decolorization ability
The purified isolates were preliminarily assessed for their dye-degrading capability using a spot inoculation method on MSM agar supplemented with Congo Red at 100 mg/L.
Each isolate was spot-inoculated onto the agar surface, while the control plate consisted of MSM agar containing Congo Red without microbial inoculation. Plates were incubated at 35 °C for 48 hours with three replicates.
The decolorization activity was evaluated by measuring the diameter (D, mm) of the clear halo zone formed around each colony, which served as an indicator of dye degradation strength
2.3. Molecular identification using 16S rRNA gene sequencing
The isolates showing the strongest qualitative decolorization activity were subjected to species-level identification using molecular methods. Total genomic DNA was extracted from bacterial biomass following the instructions of a standardized commercial extraction kit.
The 16S rRNA gene, a widely used molecular marker for bacterial taxonomy, was amplified via PCR using the universal primer pair 27F/1492R [10], with sequences:
27F: 5’-AGAGTTTGATCMTGGCTCAG-3’
1492R: 5’-GGTTACCTTGTTACGACTT-3’
PCR conditions were as follows: initial denaturation at 95 °C for 3 minutes; 30 cycles of denaturation at 95 °C for 30 seconds, annealing at 52 °C for 35 seconds, and extension at 72 °C for 1 minute; final elongation at 72 °C for 10 minutes; storage at 4 °C.
The PCR products were purified and sequenced using the Sanger method on an automated sequencing system. The obtained nucleotide sequences were analyzed using BLAST against the NCBI GenBank database to determine the highest sequence similarity, enabling accurate species and genus identification of the most efficient dye-degrading isolates.
3. RESULTS AND DISCUSSION
3.1. Isolation and qualitative screening of dye-degrading microorganisms
From the activated sludge collected at the textile wastewater treatment system of Phong Phu Corporation, the enrichment and isolation procedures yielded ten bacterial isolates capable of forming distinct clear zones on MSM agar supplemented with Congo Red. The appearance of these decolorization halos indicates that the isolates possessed the ability to either degrade or adsorb the dye to a significant extent, suggesting their potential applicability in azo dye bioremediation.
All ten isolates were identified as bacterial colonies and were designated M1 to M10 for subsequent analyses. These isolates demonstrated stable growth under the selective pressure of high dye concentration in MSM medium, which reflects their natural adaptation to the dye-rich environment of textile wastewater. The list of isolates obtained is presented in Table 1.
Table 1. Microbial isolates with azo dye decolorization capability
|
Isolate code |
Halo diameter (mm) |
Image |
Isolate code |
Halo diameter (mm) |
Image |
|
M1 |
46 |
|
M6 |
31 |
|
|
M2 |
28 |
|
M7 |
29 |
|
|
M3 |
38 |
|
M8 |
44 |
|
|
M4 |
40 |
|
M9 |
37 |
|
|
M5 |
24 |
|
M10 |
22 |
|
The qualitative screening on MSM agar supplemented with Congo Red showed that the bacterial isolates exhibited halo diameters ranging from 22 mm to 46 mm. Among them, strain M10 displayed the lowest decolorization activity, with a halo diameter of 22 mm, whereas strains M1 and M8 demonstrated the strongest decolorization capability, forming clear zones of 46 mm and 44 mm, respectively. These results indicate substantial variability in dye-degrading potential among the isolates and support the selection of the most efficient strains for further molecular identification and characterization
3.2. Molecular identification by 16S rRNA gene sequencing
The molecular identification of the dye-degrading isolates was carried out to determine their taxonomic affiliation and to better understand their potential functional roles in azo dye biodegradation. While qualitative screening on MSM agar supplemented with Congo Red provided initial evidence of decolorization capacity, species-level identification is essential to link observed phenotypes with known metabolic traits. The 16S rRNA gene, a highly conserved molecular marker routinely used in bacterial taxonomy, was selected for sequencing due to its reliability, universal applicability, and wide representation in public databases. The generated sequences were compared against the NCBI GenBank database using BLAST, allowing the determination of the closest phylogenetic relatives and the elucidation of species with documented dye-degrading or environmentally robust characteristics. This approach not only confirms the identity of the isolates but also provides scientific justification for their potential application in biodecolorization processes based on previously reported enzymatic or metabolic capabilities.
The 16S rRNA gene sequencing analysis revealed that isolate M1 shared a 99.87% sequence similarity with Bacillus subtilis, as shown in Figure 1. Bacillus subtilis is a well-documented Gram-positive bacterium commonly present in soil and wastewater environments, where it thrives under fluctuating nutrient and stress conditions. Previous studies have demonstrated that B. subtilis is capable of producing a range of oxidative enzymes, particularly laccases and peroxidases, which play an essential role in the cleavage of the azo –N=N– bond. The presence of these enzymes enables B. subtilis to catalyze both the initial reductive attack on azo dyes and the subsequent oxidation of aromatic amines formed as intermediates. This enzymatic potential suggests that isolate M1 may contribute effectively not only to dye decolorization but also to the detoxification of dye metabolites, making it a promising candidate for biological treatment of textile wastewater.
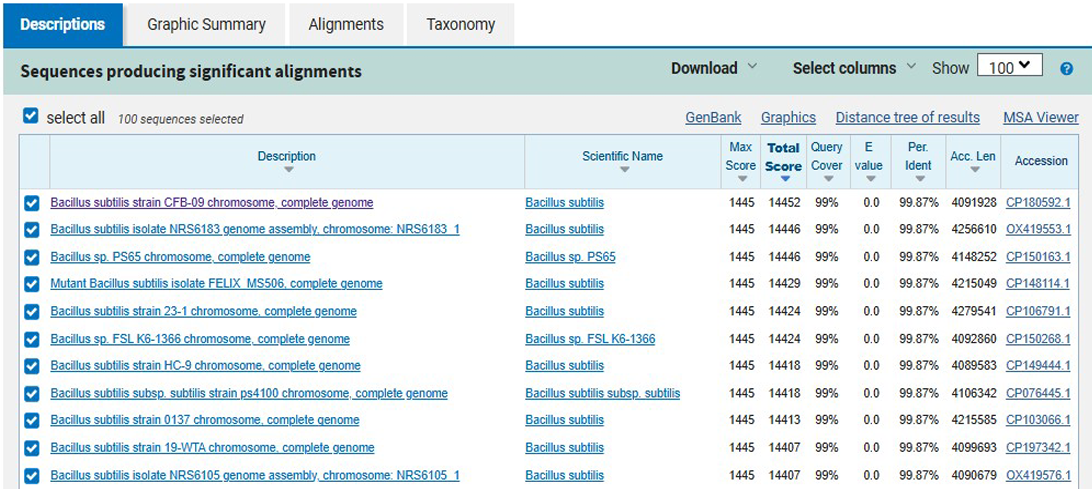
Figure 1. BLAST alignment results of the 16S rRNA gene sequence of isolate M1
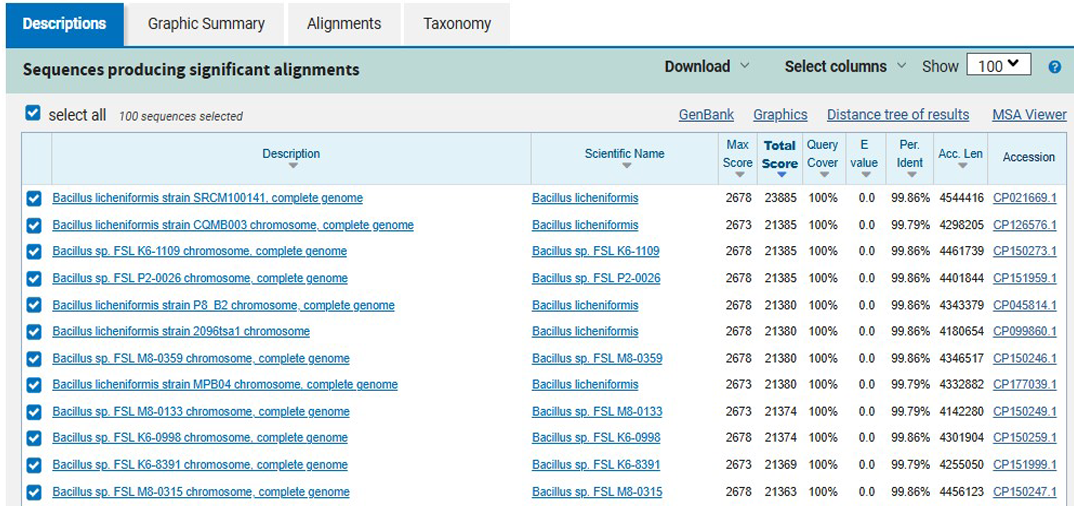
Figure 2. BLAST alignment results of the 16S rRNA gene sequence of isolate M8
Similarly, isolate M8 exhibited a 99.86% sequence similarity to Bacillus licheniformis, as shown in Figure 2. B. licheniformis, another member of the Bacillus genus, is recognized for its remarkable metabolic versatility and resilience to harsh environmental conditions, including elevated temperatures, variable pH, and high concentrations of xenobiotic compounds. This species is known to secrete abundant extracellular enzymes such as laccase, lignin peroxidase, and a broad spectrum of hydrolases, which facilitate the degradation of structurally complex dye molecules. Moreover, its robust biofilm-forming ability and tolerance to dye-induced oxidative stress further enhance its applicability in bioremediation systems subjected to high dye loads.
Taken together, the functional characteristics of B. subtilis (M1) and B. licheniformis (M8) indicate strong potential for azo dye degradation through complementary mechanisms. While both strains likely contribute to the reductive cleavage of azo bonds, B. subtilis may play a more dominant role in oxidative degradation of intermediate aromatics, whereas B. licheniformis may enhance overall dye mineralization through its diverse extracellular enzyme portfolio. The coexistence of these two species within the microbial consortium could therefore result in synergistic dye degradation and improved treatment efficiency in textile wastewater systems.
4. CONCLUSION
This study successfully isolated ten bacterial strains from the activated sludge of a textile wastewater treatment system, all of which demonstrated the ability to decolorize Congo Red with halo diameters ranging from 22 to 46 mm. Among them, strains M1 and M8 exhibited the strongest decolorization activity, forming clear zones of 46 mm and 44 mm, respectively. Based on 16S rRNA gene sequencing and BLAST analysis, strain M1 showed 99.87% similarity to Bacillus subtilis, while strain M8 exhibited 99.86% similarity to Bacillus licheniformis. Both species are well known for producing extracellular oxidative enzymes and for their high tolerance to polluted environments, suggesting strong biodegradation potential for azo dyes. These findings provide valuable insights into native dye-degrading microorganisms enriched directly from real textile wastewater treatment systems and highlight their relevance for developing specialized bioaugmentation agents. Although further quantitative assays, enzymatic profiling, and optimization studies are required, strains M1 and M8 represent promising candidates for sustainable biological treatment technologies aimed at reducing color pollution and enhancing the performance of textile wastewater treatment processes.
Trần Thị Hương 1,2 , Nguyễn Thị Nhã 3, Trần Thành 1,2,*
1 Institute of Interdisciplinary Sciences (IIS), Nguyen Tat Thanh University, Ho Chi Minh City, 700000, Vietnam.
2 Center for Hi-Tech Development, Nguyen Tat Thanh University, Saigon Hi-Tech Park, Ho Chi Minh City, 700000, Vietnam.
3 Faculty of Applied Science and Technology, Nguyen Tat Thanh University, Ho Chi Minh City, 700000, Vietnam.
(Source: The article was published on the Environment Magazine by English No. III/2025)
REFERENCES
1. N.-T. Huynh, "Status and challenges of textile and garment enterprises in Vietnam and a framework toward industry 3.5," International Journal of Logistics Research and Applications, vol. 27, no. 2, pp. 346-357, 2024.
2. H. Zollinger, Color chemistry: syntheses, properties, and applications of organic dyes and pigments. John Wiley & Sons, 2003.
3. S. I. Siddiqui et al., "Investigation of Congo red toxicity towards different living organisms: a review," Processes, vol. 11, no. 3, p. 807, 2023.
4. M. S. Nawaz and M. Ahsan, "Comparison of physico-chemical, advanced oxidation and biological techniques for the textile wastewater treatment," Alexandria Engineering Journal, vol. 53, no. 3, pp. 717-722, 2014.
5. S. S. Kumar, S. Shantkriti, T. Muruganandham, E. Murugesh, N. Rane, and S. P. Govindwar, "Bioinformatics aided microbial approach for bioremediation of wastewater containing textile dyes," Ecological Informatics, vol. 31, pp. 112-121, 2016.
6. R. A. Kristanti, M. M. F. A. Zubir, and T. Hadibarata, "Biotransformation studies of cresol red by Absidia spinosa M15," Journal of Environmental Management, vol. 172, pp. 107-111, 2016.
7. W. M. Abd El-Rahim, H. Moawad, A. Z. A. Azeiz, and M. J. Sadowsky, "Optimization of conditions for decolorization of azo-based textile dyes by multiple fungal species," Journal of biotechnology, vol. 260, pp. 11-17, 2017.
8. A. Parihar and P. Malaviya, "Textile wastewater phytoremediation using Spirodela polyrhiza (L.) Schleid. assisted by novel bacterial consortium in a two-step remediation system," Environmental Research, vol. 221, p. 115307, 2023.
9. D. L. Church, L. Cerutti, A. Gürtler, T. Griener, A. Zelazny, and S. Emler, "Performance and application of 16S rRNA gene cycle sequencing for routine identification of bacteria in the clinical microbiology laboratory," Clinical microbiology reviews, vol. 33, no. 4, pp. 10.1128/cmr. 00053-19, 2020.
10. C. Zhu, Z. Mahmood, M. S. Siddique, H. Wang, H. Anqi, and M. Sillanpää, "Structure-based long-term biodegradation of the azo dye: Insights from the bacterial community succession and efficiency comparison," Water, vol. 13, no. 21, p. 3017, 2021.